Could CRISPR have met its match with the creation of a new gene-editing tool developed at Harvard’s Wyss Institute for Biologically Inspired Engineering? Called Retron Library Recombineering or RLR for short, it may prove to be a more powerful editor capable of doing genome editing on a massive scale. The paper describing the technology states it is “a methodology for the pooled construction of mutants bearing precise genomic sequence variations and multiplex phenotypic characterization of these mutants using next-generation sequencing.”
Putting that into English requires a bit of explanation. Pooled construction refers to doing genetic modifications in volume. That is the most defining advantage RLR has over CRISPR which works as a one-off cut-and-replace gene-editing tool. Instead, RLR supports large-scale modifications of targeted genes, generating millions of mutations simultaneously.
States Max Schubert, a postdoc at the Wyss Institute, “For a long time, CRISPR was just considered a weird thing that bacteria did, and figuring out how to harness it for genome engineering changed the world. Retrons are another bacterial innovation that might also provide some important advances.”
He goes on, “RLR enabled us to do something that’s impossible to do with CRISPR: we randomly chopped up a bacterial genome, turned those genetic fragments into single-stranded DNA in situ, and used them to screen millions of sequences simultaneously.”
So what are retrons?
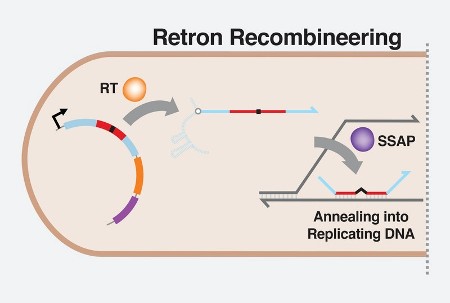
They are segments of bacterial DNA that produce fragments of single-stranded DNA (ssDNA) through reverse transcription. They have been known about for decades but the function of ssDNA escaped understanding.
With the work of the Wyss Institute researchers, it took several decades to discover how ssDNA could be used in a gene-editing process called oligonucleotide recombineering. Retrons can produce ssDNA within cells to do editing. You don’t have to come in from the outside as you do with CRISPR. As a result, the edited genes can be inherited and replicated. And CRISPR requires much greater precision. Using it can damage native DNA inadvertently making cuts to non-targeted genes. That won’t happen with ssDNA. Using RLR, therefore, researchers can perform millions of experiments simultaneously observing mutations across an entire genome. It’s no longer a one-shot deal.
The Wyss researchers don’t believe RLR will replace CRISPR. They see a combination of the two tools as a leap forward in recombinant engineering of DNA. But for now, more work is needed to improve RLR’s editing rates. The advantages are already apparent with the tool’s ability to study interactions of multiple mutations. But when combined with artificial intelligence and machine learning, RLR will allow researchers to predict future mutations.
The findings are published this week in the May 4th edition of PNAS, the Proceedings of the National Academy of Sciences of the United States of America.